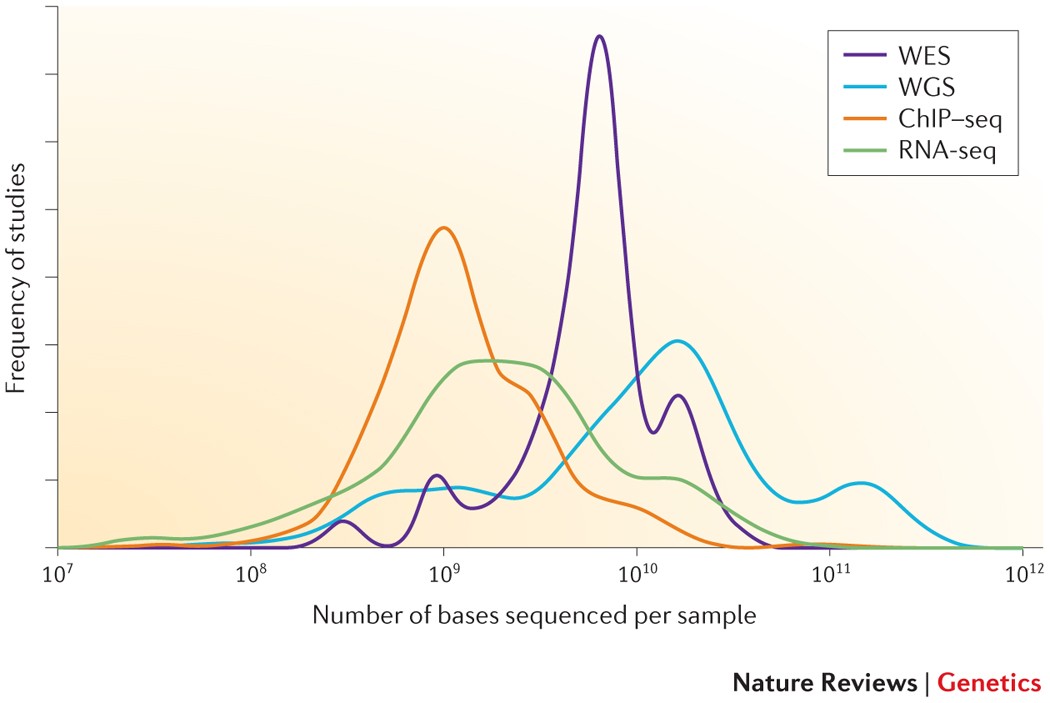
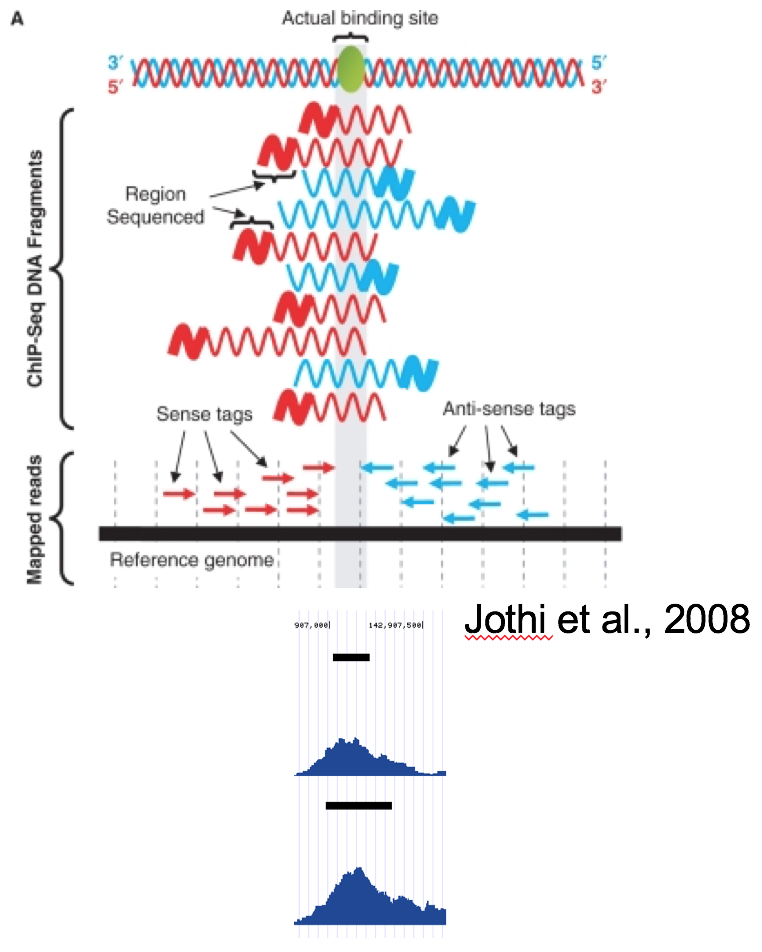
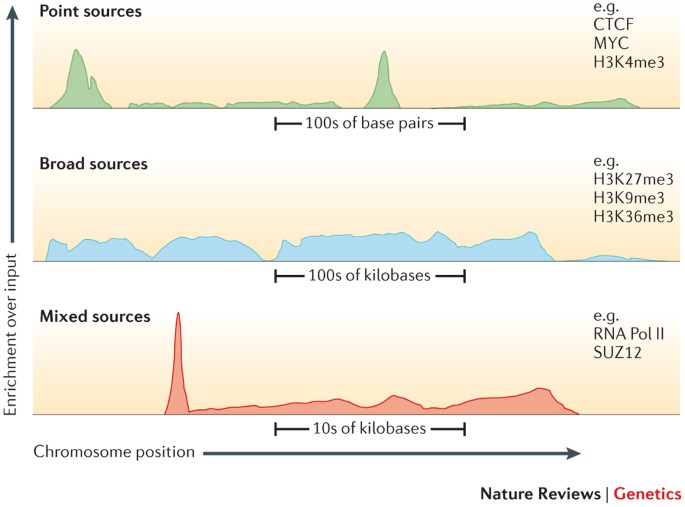
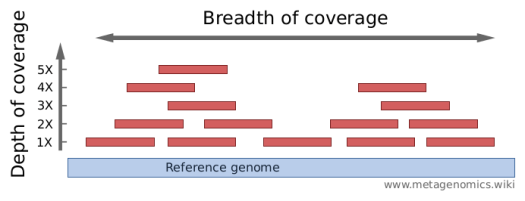
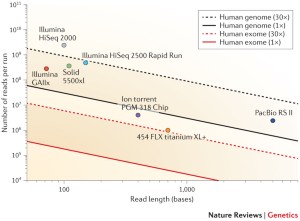
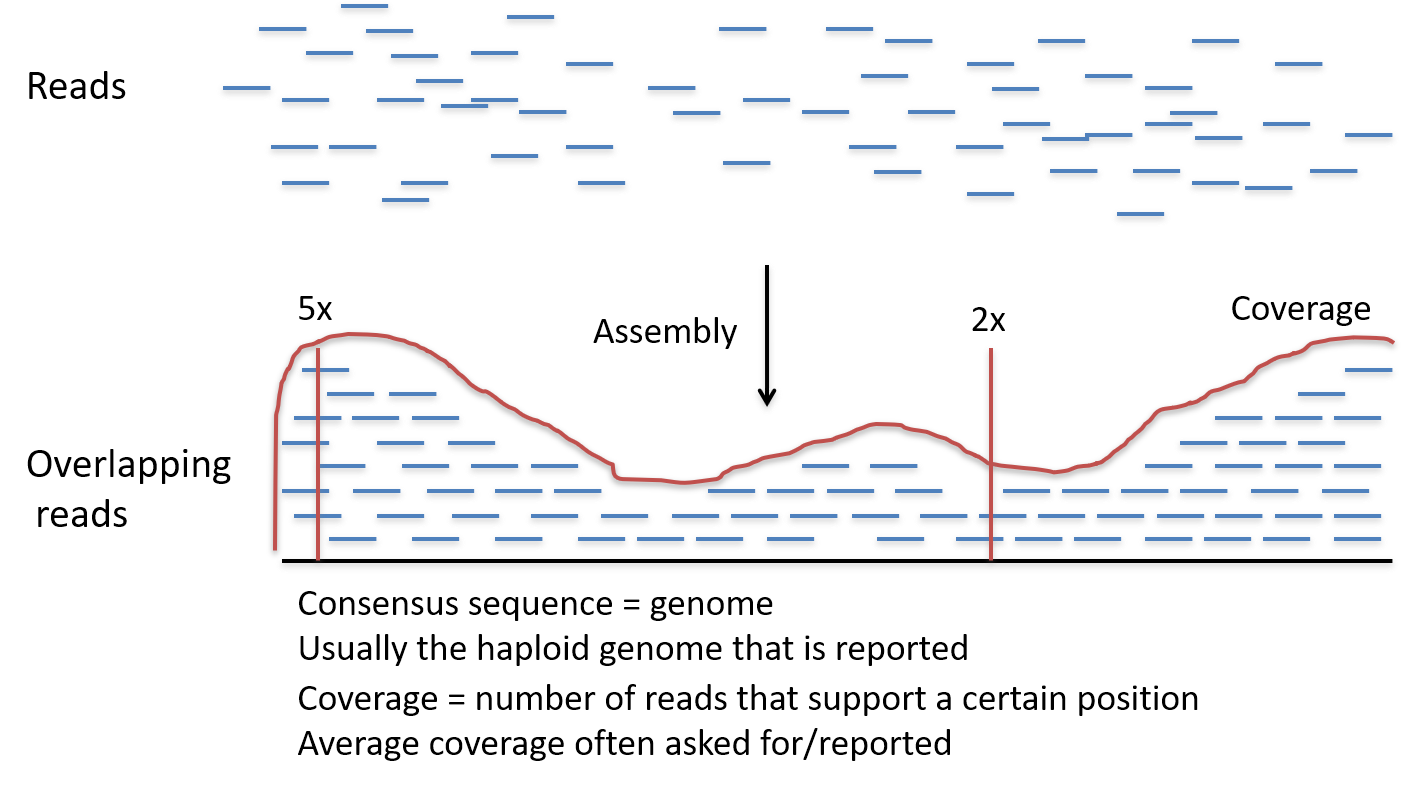
Distribution of depth-coverage. The distribution of depth-coverage of... | Download Scientific Diagram

Covcalc: Shiny App for Calculating Coverage Depth or Read Counts for Sequencing Experiments | R-bloggers
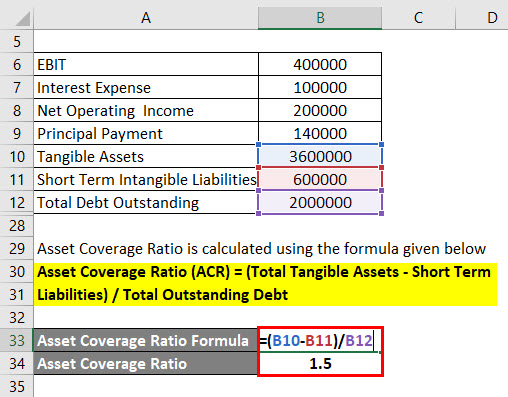
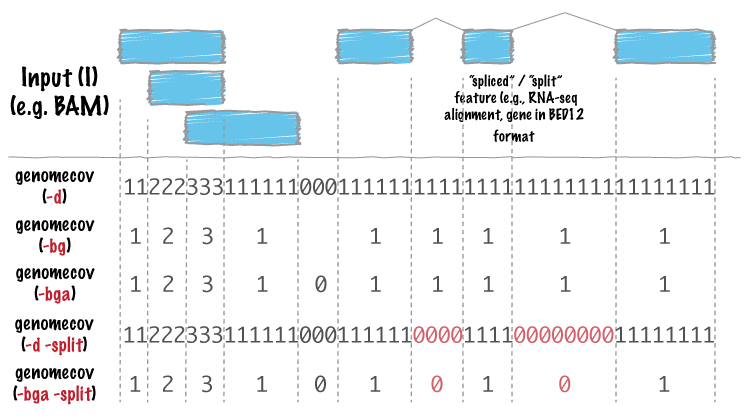
Comparison of depth of coverage calculation algorithms implemented in... | Download Scientific Diagram
GitHub - rotheconrad/00_in-situ_GeneCoverage: Workflow to calculate ANIr and sequence coverage (depth and breadth) of genome(s) / MAG(s) from metagenomes by gene, intergenic region, contig, and whole genome.